
Herbert Waldmann
Emeritus Direktor, Chemische Biologie
Research Concept
Research in the Department of Chemical Biology is focussed on the modulation and analysis of complex biological processes with approaches originating from chemistry. To this end, we have developed and employed methods for chem- and bioinformatic charting of biologically relevant chemical space, invented novel design principles and strategies for the synthesis of natural product-inspired compound collections and evaluate them in the study of biological phenomena. We employ target- and cell-based screening to find modulators of cellular processes and identify their biological targets.

In general, the major research focus of the Department is on the identification of new biologically relevant compound classes, the development of methodology for their synthesis, in particular natural product-inspired compound collections and their use in the study of biological processes. We have established a state-of-the-art screening platform, target-, cell-based and phenotypic screens and the methodology to identify the cellular targets of bioactive small molecules. The Compound Management and Screening Center (COMAS) hosts a ca. 250.000-member compound library, including ca. 15.000 proprietary in-house synthesized natural product-inspired compounds. Screening at COMAS enables access to various areas of biology, e.g. Hedgehog signaling, autophagy and unbiased biological compound characterization in multiparametric high content morphological profiling, e. g. in the Cell Painting assay.
The Chemical Genomics Centre (CGC) of the MPG established by the Department is considered a role model for successful academia/industry collaboration. As of 2018 CGC entered into a third 5-year phase and now hosts four new Research Groups funded jointly by Merck, AstraZeneca, Pfizer and the Max Planck Society.
Current Research
Natural product (NP) scaffolds represent the biologically relevant and pre-validated regions of chemical structure space explored by nature, and provide evolutionarily selected starting points for compound collection design.
We developed a novel design principle for NP-inspired compound collections which combines evolutionary selection, with rapid exploration of biologically relevant chemical space through fragment-based compound design. Unprecedented combinations and fusions of NP fragments yield to novel scaffolds that retain the chemical and biological characteristics of NPs, yet extended beyond the biologically relevant chemical space explored by nature (Figure 1). These ‘pseudo-natural products’ are not accessible through biosynthesis. Compound collections based on such molecular scaffolds may have different properties compared to NPs, and may display unprecedented biological activities. In fact, the workflow of pseudo-natural product design, synthesis and biological analysis is comparable to the concept to natural evolution such that pseudo-natural products can be regarded as the human-made equivalent, i.e. a chemical evolution of natural product structure.[1, 2]
For the preparation of pseudo-natural product libraries, we have developed novel synthetic chemistries that enable complexity-generating de novo fusion of ring systems in patterns unprecedented in nature, ideally combining ring systems that are normally not found together.
Following this design principle, we combined the scaffolds of indole- and tropane alkaloids to yield indotropanes [3], by harnessing a Cu(I)-catalysed enantioselective intermolecular 1,3-dipolar cycloaddition, and combination of the indole- and the morphine fragment delivered indomorphans.[4]
More than two fragments can be linked, for instance by combining pyrrolidine-, pyrroline- and succinimide-NP scaffolds [5], or as shown for pyrano-furo-pyridones by combining 2‑pyridone and (dihydro)pyran fragments, by means of Pd-catalysed Tsuji–Trost annulation cascades.[6] A 155‑membered chemically and biologically diverse pyrroquinoline pseudo-NP collection was generated from a few repeatedly used tetrahydro-quinoline- and pyrrolidine fragments in eight different molecular connectivities and regioisomeric arrangements. A unifying synthetic approach harnessing Ag‑catalysed 1,3-dipolar cycloadditions of azomethine ylides with electron deficient alkenes and delivered the pyrroquinoline scaffolds via Povarov-type dimerisation of enamines.[7]
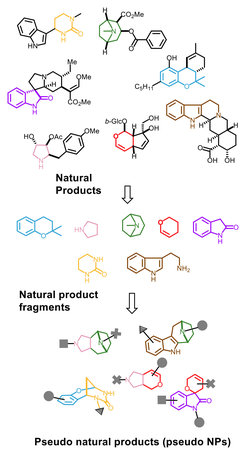
Highly NP-prevalent indole or chromanone ring systems were combined in unprecedented fusions with readily accessible NP-fragments derived from commercially available Cinchona alkaloids quinine and quinidine as well as NPs griseofulvin and sinomenine. A range of annulation reactions, including edge-fusion by indolisations and spiro-fusions by either oxa-Pictet–Spengler reactions or Kabbe condensations yielded a library of 244 chemically and biologically diverse compounds with varying ring combinatinons, connectivities, stereo-and regioisomeric fragment arrangements.[8]
Combination of the indole-containing 4H-pyranoindole fragment and the fragment-sized NP griseofulin yielded indofulvins through an iso-oxa-Pictet Spengler (IOPS) reaction as the key step to combine the two fragments with a fusion spiro connection pattern.[9] Indofulvins are novel inhibitors of starvation-induced autophagy via modulation of mitochondrial function.
Fragment-sized sesquiterpenoid lactones that are either NPs or were obtained via ring distortion were combined with the biosynthetically unrelated alkaloid fragment pyrrolidine to arrive at chemically and stereogenically diverse sesquiterpenoid alkaloid pseudo-NPs.[10]
Cheminformatic analysis of the synthesized pseudo-NP classes showed that the majority are fairly “drug like”, may have advantageous properties for molecular discovery and that their natural product likeness score resembles the score of drugs. Analysis of the ChEMBL database which hosts ca. 17 mio compounds assigned with bioactivity revealed that numerous PNPs (ca. 340.000, i.e. 20%) have been prepared during the last 40 years, probably fuelled by intuitive reasoning, and that the relative proportion of PNPs has increased steadily during this time. These findings provide independent proof-of-principle for the concept.[11]

For identification and validation of the cellular targets and the modes of action of the pseudo-NPs a suite of methods was employed, including quantitative proteomics, computational approaches, biochemical, biophysical, genetic, molecular and cell-biological techniques. Examples are shown in Figure 2.
For identification and validation of the cellular targets and the modes of action of the pseudo-NPs a suite of methods was employed, including quantitative proteomics, computational approaches, biochemical, biophysical, genetic, molecular and cell-biological techniques. Examples are shown in Figure 2.
Of particular value was the establishment of morphological profiling by means of “Cell Painting”, i.e. characterization of the cellular phenotype by selective staining of intracellular structures with five different dyes and automated microscope readout of small molecule induced perturbations. Changes in hundreds of parameters yield characteristic fingerprints that are compared with fingerprints recorded for annotated reference compounds to detect mode of action, and provide guidance if established target-ID methods fail. For instance, Cell Painting revealed that the autophagy inhibiting autoquins (for which we could not identify a putative target by means of proteomics or target prediction) are lysosomotropic agents. [12] They raise the pH and sequester Fe2+ into the lysosome leading to increased ROS production and subsequent impaired autophagosome-lysosome fusion.[12] It also allowed to qualitatively assess different biological potency and diversity of PNP classes in general,[7, 8] as well as qualitative structure-activity correlation, thereby guiding library design.[6] Even direct target identification can be achieved.[13]
Cell Painting enabled unbiased identification of bioactivity in particular in combination with other target-ID methods. For instance, combination of the method with thermal proteome profiling revealed a novel ligand of the sigma1 receptor [14], and target-independent impairment of cholesterol homeostasis by chemically different lysosome-targeting compounds.[3]
For wider coverage of NP-inspired chemical and biological space, the PNP principle was combined with macrocycle formation. PNP-macrocycles were synthesized from dimeric cinchona alkaloid-derived azomethine ylides by dipolar cycloadditions to yield pyrrolidines. The macrocyclic PNP tantalosin is a novel inducer of the lipidation of the autophagy-related LC3 protein. [15] This strategy also enabled the design and synthesis of PepNats, macrocyclic PNPs which incorporate conformation-determining NP fragments into a peptidic macrocycle [16], thereby mimicking “hot loops” in proteins involved in numerous protein-protein interactions. Biological analysis of these hybrids yielded novel and potent ligands of the SPSB2 protein that binds to iNOS, and selective ligands for AGRP-binding melanocortin (MC) receptors. [SR?]
In a more general sense macrocyclic entities like PepNats are considered “New Modalities” for modulation of novel biological targets often refractory to classical small-molecule approaches, like protein-nucleic acid interactions. [17] In extension of the PNP-derived macrocycles we have developed macrocyclic and conformationally restricted cyclic peptides in close collaboration with a research unit of Astra Zeneca, which was embedded in the Department. These include a stabilized protein tertiary structure that acts as an inhibitor of the interaction between the transcription factor TEAD and its co-repressor VGL4, that activates cell proliferation via regulation of the Hippo pathway.[18] In addition, we developed a bicyclic peptide targeting with low nanomolar affinity the interface between the scaffolding protein RbAp48 which is part of several epigenetic regulation complexes and the scaffold protein MTA1.[19]

Pseudo-NPs that induce degradation of indoleamine-2,3-dioxygenase through recruitment of a E3-ligase (“molecular glues”) were identified in a cell-based assay monitoring immune-relevant metabolism.[20] These iDegs define a novel molecular glue chemotype and the recruited E3-ligase is novel (unpublished result).
Investigation of pseudo-NPs as modulators of miRNA activity (collaboration with M. Disney, Scripps Florida) unraveled six novel RNA ligand classes which bind to numerous RNA folds and unique motifs of which 259 have not been targeted by a known small molecule binder. Transcriptome-wide mapping of bound RNA-folds showed that ca. 6% of human miRNAs are targetable by six finally triaged hits.[21] Thus, pseudo-NPs may enable targeting of nucleic acids in a wide sense.
References
- Grigalunas, M., Brakmann, S., and Waldmann, H. (2022). Chemical Evolution of Natural Product Structure. J Am Chem Soc, in the press.
- Karageorgis, G., Foley, D.J., Laraia, L., Brakmann, S., and Waldmann, H. (2021). Pseudo Natural Products-Chemical Evolution of Natural Product Structure. Angew Chem Int Ed Engl 60, 15705-15723.
- Schneidewind, T., Kapoor, S., Garivet, G., Karageorgis, G., Narayan, R., Vendrell-Navarro, G., Antonchick, A.P., Ziegler, S., and Waldmann, H. (2019). The Pseudo Natural Product Myokinasib Is a Myosin Light Chain Kinase 1 Inhibitor with Unprecedented Chemotype. Cell Chem Biol 26, 512-523 e515.
- Ceballos, J., Schwalfenberg, M., Karageorgis, G., Reckzeh, E.S., Sievers, S., Ostermann, C., Pahl, A., Sellstedt, M., Nowacki, J., Carnero Corrales, M.A., et al. (2019). Synthesis of Indomorphan Pseudo-Natural Product Inhibitors of Glucose Transporters GLUT-1 and -3. Angew Chem Int Ed Engl 58, 17016-17025.
- Akbarzadeh, M., Flegel, J., Patil, S., Shang, E., Narayan, R., Buchholzer, M., Kazemein Jasemi, N.S., Grigalunas, M., Krzyzanowski, A., Abegg, D., et al. (2022). The Pseudo-Natural Product Rhonin Targets RHOGDI.
- Christoforow, A., Wilke, J., Binici, A., Pahl, A., Ostermann, C., Sievers, S., and Waldmann, H. (2019). Design, Synthesis, and Phenotypic Profiling of Pyrano-Furo-Pyridone Pseudo Natural Products. Angew Chem Int Ed Engl 58, 14715-14723.
- Liu, J., Cremosnik, G.S., Otte, F., Pahl, A., Sievers, S., Strohmann, C., and Waldmann, H. (2021). Design, Synthesis, and Biological Evaluation of Chemically and Biologically Diverse Pyrroquinoline Pseudo Natural Products. Angew Chem Int Ed Engl 60, 4648-4656.
- Grigalunas, M., Burhop, A., Zinken, S., Pahl, A., Gally, J.M., Wild, N., Mantel, Y., Sievers, S., Foley, D.J., Scheel, R., et al. (2021). Natural product fragment combination to performance-diverse pseudo-natural products. Nat Commun 12, 1883.
- Burhop, A., Bag, S., Grigalunas, M., Woitalla, S., Bodenbinder, P., Brieger, L., Strohmann, C., Pahl, A., Sievers, S., and Waldmann, H. (2021). Synthesis of Indofulvin Pseudo-Natural Products Yields a New Autophagy Inhibitor Chemotype. Adv Sci (Weinh) 8, e2102042.
- Liu, J., Flegel, J., Otte, F., Pahl, A., Sievers, S., Strohmann, C., and Waldmann, H. (2021). Combination of Pseudo-Natural Product Design and Formal Natural Product Ring Distortion Yields Stereochemically and Biologically Diverse Pseudo-Sesquiterpenoid Alkaloids. Angew Chem Int Ed Engl 60, 21384-21395.
- Gally, J.M., Pahl, A., Czodrowski, P., and Waldmann, H. (2021). Pseudonatural Products Occur Frequently in Biologically Relevant Compounds. J Chem Inf Model 61, 5458-5468.
- Laraia, L., Garivet, G., Foley, D.J., Kaiser, N., Muller, S., Zinken, S., Pinkert, T., Wilke, J., Corkery, D., Pahl, A., et al. (2020). Image-Based Morphological Profiling Identifies a Lysosomotropic, Iron-Sequestering Autophagy Inhibitor. Angew Chem Int Ed Engl 59, 5721-5729.
- Foley, D.J., Zinken, S., Corkery, D., Laraia, L., Pahl, A., Wu, Y.W., and Waldmann, H. (2020). Phenotyping Reveals Targets of a Pseudo-Natural-Product Autophagy Inhibitor. Angew Chem Int Ed Engl 59, 12470-12476.
- 1Wilke, J., Kawamura, T., Xu, H., Brause, A., Friese, A., Metz, M., Schepmann, D., Wunsch, B., Artacho-Cordon, A., Nieto, F.R., et al. (2021). Discovery of a sigma1 receptor antagonist by combination of unbiased cell painting and thermal proteome profiling. Cell Chem Biol 28, 848-854 e845.
- Niggemeyer, G., Knyazeva, A., Gasper, R., Corkery, D., Bodenbinder, P., Holstein, J.J., Sievers, S., Wu, Y.W., and Waldmann, H. (2022). Synthesis of 20-Membered Macrocyclic Pseudo-Natural Products Yields Inducers of LC3 Lipidation. Angew Chem Int Ed Engl, e202114328.
- Gueret, S.M., Thavam, S., Carbajo, R.J., Potowski, M., Larsson, N., Dahl, G., Dellsen, A., Grossmann, T.N., Plowright, A.T., Valeur, E., et al. (2020). Macrocyclic Modalities Combining Peptide Epitopes and Natural Product Fragments. J Am Chem Soc 142, 4904-4915.
- Valeur, E., Gueret, S.M., Adihou, H., Gopalakrishnan, R., Lemurell, M., Waldmann, H., Grossmann, T.N., and Plowright, A.T. (2017). New Modalities for Challenging Targets in Drug Discovery. Angew Chem Int Ed Engl 56, 10294-10323.
- 1Adihou, H., Gopalakrishnan, R., Forster, T., Gueret, S.M., Gasper, R., Geschwindner, S., Carrillo Garcia, C., Karatas, H., Pobbati, A.V., Vazquez-Chantada, M., et al. (2020). A protein tertiary structure mimetic modulator of the Hippo signalling pathway. Nat Commun 11, 5425.
- Hart, P., Hommen, P., Noisier, A., Krzyzanowski, A., Schuler, D., Porfetye, A.T., Akbarzadeh, M., Vetter, I.R., Adihou, H., and Waldmann, H. (2021). Structure Based Design of Bicyclic Peptide Inhibitors of RbAp48. Angew Chem Int Ed Engl 60, 1813-1820.
- Hennes, E., Lampe, P., Dotsch, L., Bruning, N., Pulvermacher, L.M., Sievers, S., Ziegler, S., and Waldmann, H. (2021). Cell-Based Identification of New IDO1 Modulator Chemotypes. Angew Chem Int Ed Engl 60, 9869-9874.
- Liu, X., Tong, Y., Lee, Y., Benhamou, R.-I., Childs-Disney, J., Sievers, S., Abegg, D., Haniff, H.S., Wegner, T., Paulisch, T.O., et al. (2023). An inactive RNA-binding small molecule is rendered potently bioactive when converted into an RNA degrade. Nature, 618, 169-179
Selected references
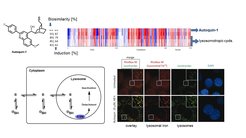
Selected Reference 1: Laraia L, Garivet G, Foley D J, Kaiser N, Müller S, Zinken S, Pinkert T, Wilke J, Corkery D, Pahl A, Sievers S, Janning P, Arenz C, Wu Y-W, Rodriguez R & Waldmann H* (2020). Image-Based Morphological Profiling Identifies a Lysosomotropic, Iron-Sequestering Autophagy Inhibitor.Angew. Chem. In.t Ed. Engl. 59, 5721-5729, doi: 10.1002/anie.201913712.
The molecular mode of action of the new autophagy inhibitor autoquin-1 was identified using image-based morphological profiling in the cell painting assay. A compound-induced fingerprint representing changes in 579 cellular parameters revealed that autoquin accumulates in lysosomes, inhibits their fusion with autophagosomes, and sequesters Fe2+ in lysosomes, resulting in an increase of lysosomal reactive oxygen species and ultimately cell death. This work demonstrates the potential of the cell painting assay to deconvolute modes of action of small molecules.
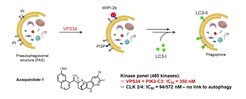
Selected Reference 2: Foley D J, Zinken S, Corkery D, Laraia L, Pahl A, Wu Y-W & Waldmann H* (2020). Phenotyping Reveals the Targets of a Pseudo-Natural Product Autophagy Inhibitor. Angew. Chem. Int. Ed. Engl. 59, 12470-12476, doi.org/10.1002/anie.202000364
The bioactivity of pseudo-NPs may be best evaluated in target agnostic cell-based assays monitoring entire cellular programs or complex phenotypes. Exploration of indocinchona alkaloid PNP bioactivities in phenotypic assays identified the novel azaquindole autophagy inhibitors. Characterization of the most potent compound, azaquindole-1, in the morphological cell painting assay, revealed that, in contrast to the parent Cinchona alkaloids, azaquindoles selectively inhibit starvation- and rapamycin-induced autophagy by targeting the lipid kinase VPS34.
Selected Reference 3: Guéret S M, Thavam S, Carbajo R J, Potowski M, Larsson N, Dahl G, Dellsén A, Grossmann T N, Plowright A T, Valeur E, Lemurell M & Waldmann H*(2020). Macrocyclic Modalities Combining Peptide Epitopes and Natural Product Fragments. J. Am. Chem. Soc.
142, 4904-4915, doi.org/10.1021/jacs.0c00269
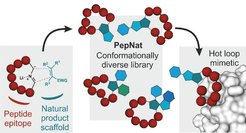
PepNats incorporate natural product (NP)-inspired structures as conformation-determining and -restricting structural elements into macrocyclic hot loop-derived peptides. Macrocyclic PepNats representing hot loops of inducible nitric oxide synthase (iNOS) and human agouti-related protein (AGRP) yielded novel and potent ligands of the SPRY domain-containing SOCS box protein 2 (SPSB2) that binds to iNOS, and selective ligands for AGRP-binding melanocortin (MC) receptors. NP-inspired fragment absolute configuration determines the conformation of the peptide part responsible for binding. These results demonstrate that combination of NP-inspired scaffolds with peptidic epitopes enables identification of novel hot loop mimics.
Selected Reference 4: Adihou H, Gopalakrishnan R, Förster T, Guéret S M, Gasper R, Geschwindner S, Carrillo García C, Karatas A, Pobbati A V, Vazquez‐Chantada M, Davey P, Wassvik C M, Sheng Pang J K, Seng Soh B, Hong W, Chiarparin E, Schade E, Plowright A T, Valeur E, Lemurell M, Grossmann T N & Waldmann H* (2020). A protein tertiary structure mimetic modulator of the Hippo signalling pathway Nature Commun. 11:5425, https://doi.org/10.1038/s41467-020-19224-8
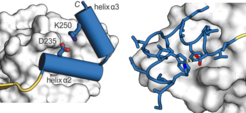
Chemical modulation of transcription factor activity is hampered by the difficulties associated with the targeting of PPIs, in particular when extended and flat protein interfaces are involved. The mimicry of a single secondary structure element may be insufficient to obtain high binding affinities. We have designed a stabilized protein tertiary structure that acts as an inhibitor of the interaction between the transcription factor TEAD and its co-repressor VGL4, both playing a central role in the Hippo signalling pathway. Modification of the inhibitor with a cell-penetrating entity yielded a cell-permeable proteomimetic that activates cell proliferation via regulation of the Hippo pathway, highlighting the potential of protein tertiary structure mimetics as an emerging class of PPI modulators.
Selected Reference 5: Liu J, Cremosnik G S, Otte F, Pahl A, Sievers S, Strohmann C & Waldmann H* (2021). Design, Synthesis,and Biological Evaluation of Chemically and Biologically Diverse Pyrroquinoline Pseudo Natural Products Angew. Chem. Int. Ed. Engl. 60, 4648 – 4656, doi.org/10.1002/anie.202013731
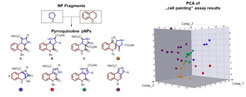
The paper describes the design and synthesis of a 155-member pyrroquinoline pseudo-NP collection in which fragments characteristic of the tetrahydroquinoline and pyrrolidine NP classes are combined with eight different connectivities and regioisomeric arrangements. Cheminformatic analysis and biological evaluation of the compound collection by means of phenotyping in the morphological “cell painting” assay followed by principal component analysis revealed that the pseudo-NP classes are chemically diverse and that bioactivity patterns differ markedly, and are dependent on connectivity and regioisomeric arrangement of the fragments.
Selected Reference 6: Grigalunas M, Burhop A, Zinken S, Pahl A, Gally J-M, Wild N, Mantel Y, Sievers S, Foley D J, Scheel R, Strohmann C, Antonchick A P & Waldmann H* (2021). Natural Product Fragment Combination to Performance-Diverse Pseudo-Natural Products Nat. Commun. 12:1883; https://doi.org/10.1038/s41467-021-22174-4
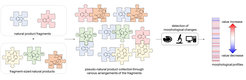
Natural product structure and fragment-based compound development inspire pseudo-natural product design. We describe the synthetic combination of the fragment-sized natural products quinine, quinidine, sinomenine, and griseofulvin with chromanone or indole-containing fragments to provide a 244-member pseudo-natural product collection. The resulting eight pseudo-natural product classes are chemically diverse and share both drug- and natural product-like properties. Unbiased biological evaluation by cell painting demonstrates that bioactivity of pseudo-natural products, guiding natural products, and fragments differ. Identification of phenotypic fragment dominance enables design of compound classes with correctly predicted bioactivity.
Selected Reference 7: Schneidewind T, Brause A, Schölermann B, Sievers S, Pahl A, Sankar M G, Winzker M, Janning P, Kumar K, Ziegler S & Waldmann H* (2021). Combined morphological and proteome profiling reveals target-independent impairment of cholesterol homeostasis. Cell Chemical Biology 28, 1780–1794; https://doi.org/10.1016/j.chembiol.2021.06.003

Unbiased profiling approaches generate a holistic view of bioactivity space and may enable identification of non-protein targets. We report the identification of a new, large bioactivity cluster comprised of numerous well characterized drugs with very different primary targets using a combination of the morphological Cell Painting Assay and proteome profiling. Cluster members alter cholesterol homeostasis and localization due to their physicochemical properties that lead to protonation and accumulation in lysosomes, an increase in lysosomal pH and a disturbed cholesterol homeostasis.
Selected Reference 8: Wilke J, Kawamura T, Xu H, Brause A, Friese A, Metz M, Schepmann D, Wünsch B, Artacho-Cordón A, Nieto F R, Watanabe N, Osada H, Ziegler S & Waldmann H* (2021). Discovery of a novel σ1 receptor antagonist by combination of unbiased Cell Painting and thermal proteome profiling. Cell Chemical Biology 28, 848–854; doi.org/10.1016/j.chembiol.2021.01.009
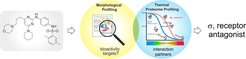
Unbiased cellular profiling approaches record hundreds of parameters upon compound perturbation to map bioactivity in a broad biological context and may link a profile to the molecular target or mode of action. The paper reports the discovery of the diaminopyrimidine DP68 as a Sigma 1 (s1) receptor antagonist by combining morphological profiling using the Cell Painting assay and thermal proteome profiling. Our results highlight that integration of complementary profiling approaches may enable both detection of bioactivity and target identification for small molecules.
Selected Reference 9: Akbarzadeh M, Flegel J, Patil S, Shang E, Narayan R, Buchholzer M, Kazemein Jasemi N S, Grigalunas M, Krzyzanowski A, Abegg D, Shuster A, Potowski M, Karatas H, Karageorgis G, Mosaddeghzadeh N, Zischinsky M-L, Merten C, Golz C, Brieger L, Strohmann C, Antonchick A P, Janning P, Adibekian A, Goody R S, Ahmadian M R, Ziegler S & Waldmann H* (2022). The Pseudo-Natural Product Rhonin Targets RHOGDI. Angew. Chem. Int. Ed. Engl., 61, e202115193; doi.org/10.1002/anie.202115193
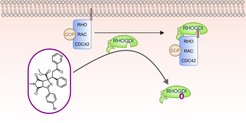
We describe the de novo combination of different 5-membered NP-derived N-heteroatom fragments to structurally unprecedented pseudo-NPs in an efficient complexity-generating and enantioselective one-pot synthesis sequence. Investigation of the pseudo-NPs in unbiased phenotypic assays and target identification led to the discovery of the first small-molecule ligand of the RHO GDP-dissociation inhibitor 1 (RHOGDI1), termed Rhonin. Rhonin inhibits the binding of the RHOGDI1 chaperone to GDP-bound RHO GTPases and alters the subcellular localization of RHO GTPases.
Selected Reference 10: Liu X, Tong Y, Lee Y, Benhamou R I, Childs-Disney J, Sievers S, Grefe M, Crynen G, Costales M G, Abegg D, Haniff H S, Wegner T, Paulisch T O, Lekah E, Adibekian A, Glorius F, Waldmann H & Disney M D (2023). An inactive RNA-binding small molecule is rendered potently bioactive when converted into an RNA degrader. Nature, 618, 169-179

For the discovery of small molecules that bind RNA structures selectively a natural-product inspired small molecule collection and three dimensionally folded RNA structures were investigated, probing >61 million interactions. Six new chemotypes that avidly bind RNA and ~2,000 new RNA folds that bind small molecules were discovered and mined across the human transcriptome to define targetable structures and thus structure-activity relationships for the small molecules and RNA folds transcriptome-wide. For modulation of RNA biology by cleaving the target via a ribonuclease targeting chimera, the substrate specificity for RNase L was overlaid with the binding landscape of the new RNA binders to yield novel degraders, i.e. of microRNA-155 (pre-miR-155) in multiple cell lines and disease settings. These studies illustrate the advantageous properties of natural predict derived RNA ligands that can be converted into potent and specific modulators of RNA function.













