Ingrid Vetter
Projektgruppenleiterin, Mechanistische Zellbiologie
Strukturaufklärung von Proteinen und Proteinkomplexen
Research Concept
Our research area is the determination of the three-dimensional structure of proteins and protein complexes via X-ray Crystallography. We are interested especially in nuclear transport proteins and their associates and investigate protein-protein interactions with respect to nuclear transport/signal transduction mechanisms. In addition, we investigate proteins of the nuclear pore itself with the future goal to elucidate the structure of the pore scaffold.
Nuclear transport is one of the most important activities in eukaryotic cells. The essential regulator of nuclear transport is the small GTP binding protein Ran that determines the directionality of the transport process via the phosphorylation state of the bound nucleotide. The interaction of Ran with import receptors is quite well understood, but its interactions with export receptors and with the proteins of the nuclear pore, the nucleoporins (NUPs), are less well characterized.
Current Research
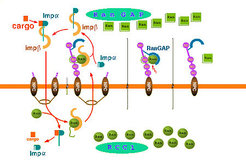
One Cycle of Protein Import is Depicted Schematically in the Following Figure of the right Handside:

Ran binding proteins (RanBP) are essential proteins involved in dissociation of nuclear transport complexes in the cytoplasm. Our structure of the complex of Ran with RanGAP and RanBP1 and the detailed biochemical investigations revealed the mechanism by which RanBP1 renders the GTPase reaction more efficient. The function of Ran-binding proteins in interaction with import receptor complexes is quite well investigated, but much less is known e.g. about the function of Ran binding proteins in export complexes.
We are characterizing the function of several Ran binding domains and are trying to crystallize them in complex with import/export receptors.
Based on the crystal structures and the biochemical and structural evidences, some steps of nuclear transport have been visualized with the software "MAYA" to get an impression of the nuclear transport process. The import receptor Importin beta is shown in yellow, Importin alpha in cyan, a transport substrate (in this case NF-kappaB) in orange, Ran in green, RanBD (in the cytoplasmic filament) in red and RanGAP (at the tip of the cytoplasmic filament) in cyan. The model of the pore has been built according to the electron microscopy studies of N. Pante and U. Aebi.
The Nuclear Pore Complex (NPC)
The nuclear pore complex (NPC) is the most complex transport system in eukaryotic cells. In spite of the research effort of the last few years, the atomic structure of the NPC and the details of the transport mechanism through the nuclear envelope are still unknown. The complete nuclear pore of yeast consists of only approx. 30 nucleoporins (NUPs) that are arranged in an 8-fold symmetrical manner, forming a complex of approx 60 MDa in yeast (NPC). Localization and interaction studies revealed the position of individual NUPs within the NPC, and several major subcomplexes could be identified. Most of the NUPs associate with and dissociate from the nuclear pore in a highly dynamic manner, but the some subcomplexes are found to be stably associated with the nuclear pore and assumed to build the central scaffold of the pore itself.
In collaboration with the group E. Hurt (BZH, Heidelberg) and the group of
A. Wittinghofer (MPI, Dortmund) structures of selected nucleoporins will be determined with the aid of X-ray crystallography, if possible, in complexes with the GTP-binding protein Ran and transport factors.
Since most NUPs are very large proteins (100-200 kDa) and determining crystallizable fragments would have been a major bottleneck for structure determination, we started the development of a new DNA-based high-throughput method to identify soluble yeast NUP domains using a GFP folding assay. This method should overcome the problem that Nup domain boundaries cannot be predicted easily due to weak sequence homologies.
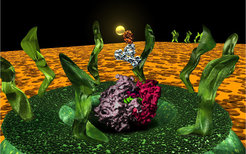
The picture on the right handside shows a model of the nuclear pore together with the atomic structure of a ribosome (pink/violet) and an antibody labeled with a 5nm gold particle to show the size relationship.
Instrumentation
The crystallography facilities were greatly expanded and modernized in 2004 and 2005. A new top-of-the-line Rigaku X-ray generator equipped with two MAR345 desktop beamlines, one with a robotic sample changer, allows efficient in-house testing of small crystals, thus making it easier to use synchrotron beamtime efficiently. A Mosquito crystallization robot is used for fast screening with protein drop volumes as low as 50 nl, and a Zinsser Lissy robot is available for pipetting of masterblocks and custom screens.




