
Andrea Musacchio
Direktor, Mechanistische Zellbiologie
Research Concept
The molecular processes of cell division
Enchantment with holistic approaches has not diminished the paramount importance that detailed investigations of molecular mechanisms have for the advancement of Biology. Life at “machine level” is complex and full of wonders. For instance, ribosomes consist of tens of polypeptides and several RNAs working seamlessly together to decode RNA and translate its code into protein products. The essence of the processes controlled by cellular molecular machines is almost invariably dynamic. Stable scaffolds representing the structural cores of molecular machines harness energy to achieve complex regulation, interacting dynamically with associated factors in cycles that can repeat themselves with frequencies as high as hundred Hertz or more. Understanding the complex chemistry of life is the only safe guide to meaningful biological perturbation experiments, and is equally invaluable for the interpretation of the perturbations that occur in disease.
Current Research
The organization and function of kinetochores
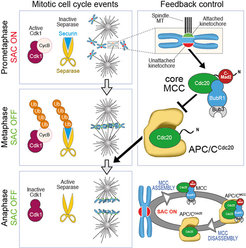
In the last 15 years, our group applied this tenet to the study of the organization and function of kinetochores. Kinetochores are multi-subunit protein complexes known to perform three main functions during chromosome segregation in mitosis and meiosis. First, kinetochores create physical linkages between the replicated chromosomes (sister chromatids) and microtubules of the mitotic spindle, so to allow transport of the sisters to the spindle poles during anaphase.
Second, kinetochores not only create the linkages, but also distinguish those that support sister bi-orientation (the correct configuration for accurate chromosome segregation) from those that don’t, and promote selective elimination of faulty attachment (a process named error correction). Finally, kinetochores host and regulate a cell cycle checkpoint, the spindle assembly checkpoint (SAC), which coordinates chromosome alignment with mitotic exit (Figure 1).
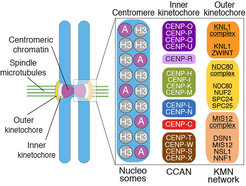
Kinetochores are built on a specialized chromosome locus named the centromere, a specialized chromatin domain hosting a histone H3 variant named CENP-A (Figure 2). CENP-A is held responsible for the “epigenetic” specification of centromeres, i.e. the ability of centromeres to propagate themselves through generations without generally resorting to specialized genetic determinants (i.e. specific DNA sequences). Studies in the early to mid 2000s led to the discovery of a number of core kinetochore subunits, including several subunits associated with the CENP-A nucleosome and responsible for the deposition of new CENP-A at every new cell cycle, as well as subunits involved in microtubule binding and SAC control [1, 2]. The structural core of the kinetochore is made of ~30 different subunits, but the overall count of proteins estimated to reside at kinetochores during mitosis is in the order of ~100, numbers that identify kinetochores as one of the largest known macromolecular complexes in the cell. The study of kinetochores is therefore a significant challenge.
Biochemical reconstitution of the kinetochore
Contrarily to other widely studied macromolecular machines, such as ribosomes or the proteasome, kinetochores cannot be easily isolated in a native form, not least because their association with centromeric chromatin complicates their physical isolation from chromosomes. Even if recent progress on the isolation of the simplest kinetochores, those of S. cerevisiae, has been reported [3], our studies on the more complex human kinetochores have relied on biochemical reconstitution with recombinant proteins, which we deem likely to lead to more homogeneous and tractable preparations.
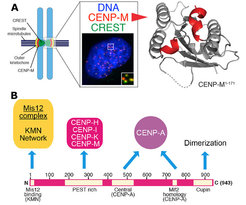
In a three-pronged approach, we complemented biochemical reconstitution with structural analyses and cellular investigations, in an attempt to reach, in the long run, a comprehensive view of kinetochore structure and functional dynamics (Figure 3). Reconstitution and structural analysis of CENP-M (Basilico et al. 2014 , Figure1), of the Bub3:Bub1:MELT complex (Primorac et al. 2013) , of the Mis12 complex (Petrovic et al. 2014, Klare et al. 2015 Figure 3B) are highlights of our recent structural work.
Our early work focused on individual subunits or sub-complexes of the SAC and of kinetochores, such as the Mad1:Mad2 SAC complex or the Ndc80 complex [4-6], but our ambition has grown with time. After reconstituting an “outer kinetochore” supra-molecular complex of the Knl1 complex, Mis12 complex, Ndc80 complex (KMN) network [7, 8], which is involved in microtubule binding and SAC recruitment (Figure 2), we started addressing the organization of the inner kinetochore (i.e. the kinetochore domain proximal to the CENP-A chromatin [1]. In three studies, we identified physical interactions between proteins in the constitutive centromere-associated network (CCAN, Figure 2, 3), a group of 16 proteins known to be closely associated with CENP-A, and also identified a crucial linkage between the CCAN and KMN complexes [9-11]. The picture emerging from our work and from that of other scholars is that the CCAN consists of sub-complexes endowed with significant autonomous stability (Basilico et al. 2014, Klare et al. 2015, Weir et al. 2016). The reconstitution of the 4-subunit CENP-HIKM complex was instrumental to support further developments [10] (comprehensive view of kinetochore structure and functional dynamics (Figure 3).
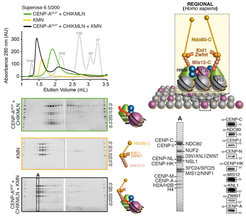
With the availability of high-grade recombinant versions of essentially all core kinetochore components, we attempted the reconstitution of kinetochore particles of near physiologic complexity. The study by Weir et al. (2016) reports the reconstitution of a 21-subunit kinetochore particle containing the CENP-A nucleosome, 7 core CCAN subunits (CENP-CHIKMLN), and 10 KMN subunits (Figure 4). The reconstitution reveals important concepts of kinetochore architecture and function. For instance, incorporation of two CENP-A binding activities like CENP-C and CENP-LN in the same complex (the CENP-CHIKMLN complex) promotes a very significant increase in CENP-A binding affinity, reflecting binding cooperativity. This mechanism explains how CCAN interacts selectively with CENP-A in comparison to canonical H3 nucleosomes. We are currently extending the reconstitution to additional CCAN components, including the CENP-T:CENP:W:CENP-S:CENP-X complex (CENP-TWSX) and the CENP-O:CENP:P:CENP:Q:CENP:R:CENP-U complex (CENP-OPRQU). The TWSX complex contributes to the multiplication of the number of outer kinetochore KMN subunits [12-15]. In summary, we have identified conditions for the reconstitution of a 30-subunit kinetochore core complex, including the 4-subunit nucleosome, the 16-subunit CCAN complex, and the 10-subunit KMN network (Figure 2).
Literature
1 Fukagawa, T., and Earnshaw, W.C. (2014). The centromere: chromatin foundation for the kinetochore machinery. Developmental cell 30, 496-508.
2. Musacchio, A. (2015). The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Current biology : CB 25, R1002-1018.
3. Akiyoshi, B., Sarangapani, K.K., Powers, A.F., Nelson, C.R., Reichow, S.L., Arellano-Santoyo, H., Gonen, T., Ranish, J.A., Asbury, C.L., and Biggins, S. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468, 576-579.
4. Ciferri, C., Pasqualato, S., Screpanti, E., Varetti, G., Santaguida, S., Dos Reis, G., Maiolica, A., Polka, J., De Luca, J.G., De Wulf, P., et al. (2008). Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427-439.
5. De Antoni, A., Pearson, C.G., Cimini, D., Canman, J.C., Sala, V., Nezi, L., Mapelli, M., Sironi, L., Faretta, M., Salmon, E.D., et al. (2005). The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Current biology : CB 15, 214-225.
6. Sironi, L., Mapelli, M., Knapp, S., De Antoni, A., Jeang, K.T., and Musacchio, A. (2002). Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. The EMBO journal 21, 2496-2506.
7. Petrovic, A., Mosalaganti, S., Keller, J., Mattiuzzo, M., Overlack, K., Krenn, V., De Antoni, A., Wohlgemuth, S., Cecatiello, V., Pasqualato, S., et al. (2014). Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Molecular cell 53, 591-605.
8. Petrovic, A., Pasqualato, S., Dube, P., Krenn, V., Santaguida, S., Cittaro, D., Monzani, S., Massimiliano, L., Keller, J., Tarricone, A., et al. (2010). The MIS12 complex is a protein interaction hub for outer kinetochore assembly. The Journal of cell biology 190, 835-852.
9. Klare, K., Weir, J.R., Basilico, F., Zimniak, T., Massimiliano, L., Ludwigs, N., Herzog, F., and Musacchio, A. (2015). CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. The Journal of cell biology 210, 11-22.
10. Basilico, F., Maffini, S., Weir, J.R., Prumbaum, D., Rojas, A.M., Zimniak, T., De Antoni, A., Jeganathan, S., Voss, B., van Gerwen, S., et al. (2014). The pseudo GTPase CENP-M drives human kinetochore assembly. Elife 3, e02978.
11. Screpanti, E., De Antoni, A., Alushin, G.M., Petrovic, A., Melis, T., Nogales, E., and Musacchio, A. (2011). Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Current biology : CB 21, 391-398.
12. Schleiffer, A., Maier, M., Litos, G., Lampert, F., Hornung, P., Mechtler, K., and Westermann, S. (2012). CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nature cell biology 14, 604-613.
13. Gascoigne, K.E., Takeuchi, K., Suzuki, A., Hori, T., Fukagawa, T., and Cheeseman, I.M. (2011). Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410-422.
14. Rago, F., Gascoigne, K.E., and Cheeseman, I.M. (2015). Distinct Organization and Regulation of the Outer Kinetochore KMN Network Downstream of CENP-C and CENP-T. Current biology : CB 25, 671-677.
15. Kim, S., and Yu, H. (2015). Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. The Journal of cell biology 208, 181-196.



