
Peter Bieling
Research Group Leader, Systemic Cell Biology
Regulatory mechanisms of cell morphology
Research Concept
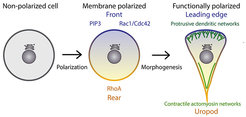
We are interested in the molecular mechanisms that regulate changes in cellular morphology and the underlying polarization of the signaling molecules that control these processes. Nearly all cells display some form of polarity, which allows them to perform spatially complex functions such as motility or the formation of tissues and organs. Polarization requires a complex interplay between biochemical signals that are generated at the plasma membrane and cytoplasmic molecules, most importantly the actin cytoskeleton. In eukaryotic organisms, cellular symmetry is initially broken through the integration of signaling by membrane associated receptors (e.g. GPCRs and RTKs). Information is then relayed to downstream effectors, which regulate the synthesis of phosphatidylinositols (PtdIns) lipids and the activation of Rho-family GTPases. These two intimately linked classes of signaling systems synergistically control cell shape, organization and movement by guiding the assembly of functionally distinct actin structures (Figure 1).
While many key players of cell polarity and morphogenesis are now known, we currently do not understand how spatial signaling systems can break symmetry and how the size and shape of their emerging domains is determined. Importantly, we have not yet managed to re-built systems from defined components that recapitulate membrane polarization in vitro. Furthermore, we do not understand the molecular mechanisms by which PtdIns and Rho-family GTPases intersect. Finally, how the combinatorial action of these spatial control networks organizes the actin cytoskeleton to control cell shape and generate movement is unclear.
Using cell motility as a testbed, we plan to answer these fundamental questions by using minimal in vitro systems. We test our understanding of these systems by employing synthetic biology techniques to rationally engineer biomimetic networks capable of autonomously breaking membrane symmetry. Instead of studying membrane polarity and actin assembly in their complex cellular environment, we want to reconstitute these processes from purified components using a bottom-up approach. We combine multi-protein reconstitution with advanced fluorescence imaging (TIRFM, FLIM) down to the level of single molecules. This allows us to study and manipulate all biochemical activities in great detail to reveal the design principles underlying self-organization of signaling networks.
Current Research
Reconstitution of self-organized membrane polarity
Phosphatidylinositols and Rho-GTPases are the key components at the plasma membrane that act as a compass for directing the spatiotemporal regulation of the actin cytoskeleton. Cells utilize complex networks of regulatory proteins, which include kinases, phosphatases, GEFs and GAPs to polarize and control the distribution and activity of these signaling molecules. Although many network components have been identified, the minimal biochemical system(s) required for establishment and maintenance of polarity remains unclear.
Computational models of polarization predict that positive and negative feedback loops are essential regulatory links for spatial symmetry breaking of membrane associate reaction diffusion systems. Can we employ these design principles to engineer systems capable of autonomously generating membrane polarity? In collaboration with Scott Hansen and Jay Groves (UC Berkeley), we are biochemically reconstituting simple reaction diffusion systems that can drive self-organization of PtdIns lipids on lipid bilayers. Our goal is to establish simple PtdIns based polarizers that can function as a spatial cue for directing actin assembly on model membranes in vitro.
Ptdins- and rho gtpase-mediated control of actin organization and dynamics
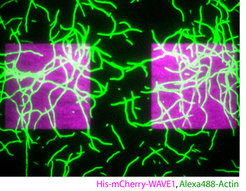
Local control of actin dynamics by PtdIns and Rho-family GTPases is likely key, since many prominent actin regulators are directly and often synergistically regulated by these two signaling modules. However, we do not understand the systems-level feedback loops that coordinate membrane signaling and actin assembly in space and time. To this aim, we are studying protrusive and contractile actin structures in the context of the membrane-associated molecules that control their assembly.
Membrane polarity and actin assembly in cell-like geometries
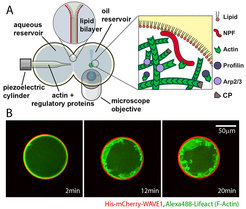
We know that the cytoskeleton sculptures the cell in response to spatial signals generated at the plasma membrane. Whether and how cell shape also feeds back on these biochemical systems, however, is far less appreciated. Theory predicts that self-organized patterning systems will strongly be influenced by geometric boundary conditions. Moreover, confinement has the potential to control biochemistry locally through depletion effects.
We are therefore developing assays to build actin networks on the inner membrane of giant unilamellar vesicles (GUVs). These micro-compartments mimic the boundary conditions and volume-to-surface ratios typically found in living cells. These experiments provide key insight into how geometry directly governs membrane polarity and shape changes.


