Cryo-EM structure of the ClpXP protein degradation machinery
Raisch T, Chang CT, Levdansky Y, Muthukumar S, Gatsogiannis C, Balogh D, Merino F, Sieber S, Raunser S (2019). Nat. Struct. Mol. Biol.
doi: 10.1038/s41594-019-0304-0.
Antibiotics are still the most important weapon to fight bacterial infections. However, medicine is running out of ammunition due to increasing resistance. One approach to the development of innovative antibiotics aims at the degradation process of defective proteins in bacteria.
The group of Prof. Dr. Stephan Sieber (Technical University of Munich) together with the group of Prof. Dr. Stefan Raunser, Director at the Max Planck Institute (MPI) of Molecular Physiology in Dortmund, have now unveiled the first high-resolution 3D structure of the protein degrading complex ClpX-ClpP and thus created an important structural basis for future pharmacological applications.
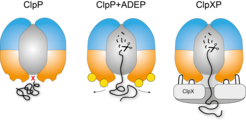
Regulation of ClpP by ClpX and ADEP. Left: The central pore of the ClpP protease is closed and entry of folded proteins into the proteolytic chamber is not allowed. Middle: ADEP binding to the binding pockets of ClpP induces pore opening. The proteolytic chamber is now accessible for unfolded proteins, leading to unregulated protein degradation and cell death. Right: ClpX binds in the same hydrophobic pockets on ClpP but does not induce pore opening. ClpP and ClpX form a continuous pore instead, with ClpX unfolding target proteins and forwarding them to the proteolytic chamber of ClpP for degradation in a regulated manner.
MPI Dortmund
